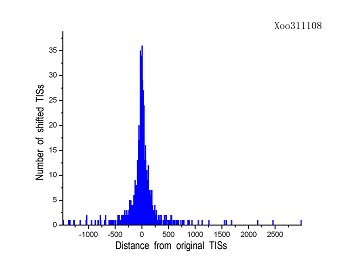
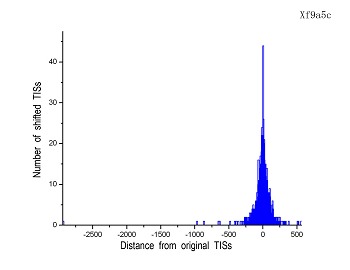
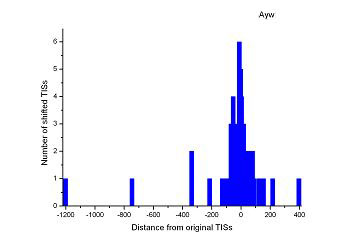
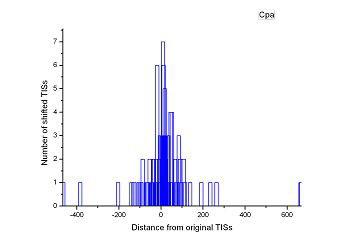
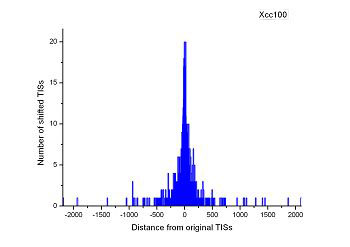
Figure 4 (a-p). The histogram for the shift length of translation initiation sites (TISs) for the 28 plant pathogens. Negative and positive values indicate the length of 5กฏ-shift and 3กฏ-shift from the RefSeq annotation, respectively.
 |
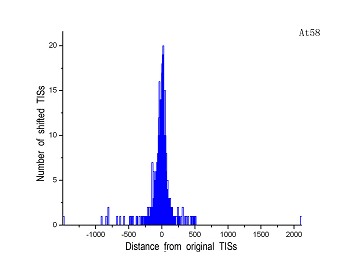 |
| (1) Acidovorax avenae subsp. citrulli AAC00-1 | (2) Agrobacterium tumefaciens str. C58 |
 |
 |
| (3) Clavibacter michiganensis subsp. michiganensis NCPPB 382 | (4) Erwinia carotovora subsp. atroseptica SCRI1043 |
 |
 |
| (5) Pseudomonas syringae pv. phaseolicola 1448A | (6) Pseudomonas syringae pv. syringae B728a |
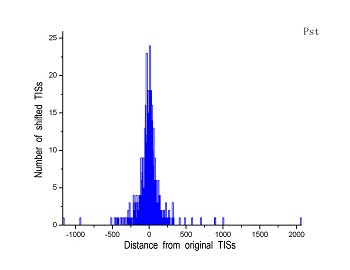 |
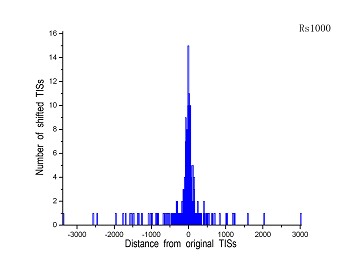 |
| (7) Pseudomonas syringae pv. tomato str.DC3000 | (8) Ralstonia solanacearum GMI1000 |
 |
 |
| (9) Xanthomonas axonopodis pv. citri str. 306 | (10) Xanthomonas campestris pv. campestris str. 8004 |
 |
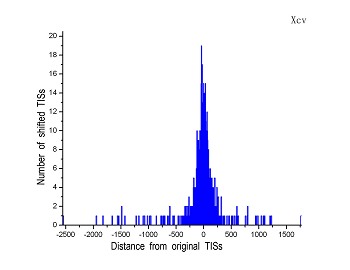 |
| (11) Xanthomonas campestris pv. campestris str. ATCC 33913 | (12) Xanthomonas campestris pv. vesicatoria str. 85-10 |
 |
 |
| (13) Xanthomonas oryzae pv. oryzae MAFF 311018 | (14) Xanthomonas oryzae pv. oryzae KACC10331 |
 |
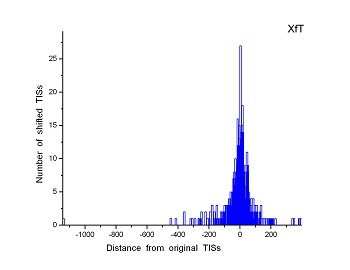 |
| (15) Xylella fastidiosa 9a5c | (16) Xylella fastidiosa Temecula1 |
 |
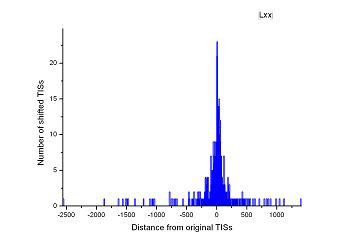 |
| (17) Aster yellows witches'-broom phytoplasma strain AY-WB | (18) Leifsonia xyli subsp. xyli str. CTCB07 |
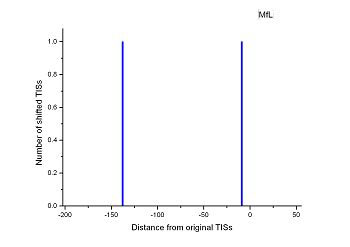 |
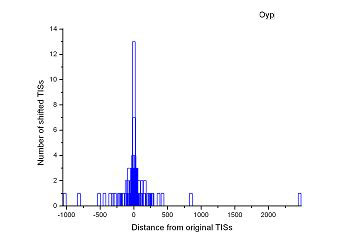 |
| (19) Mesoplasma florum L1 | (20) Onion yellows phytoplasma OY-M |
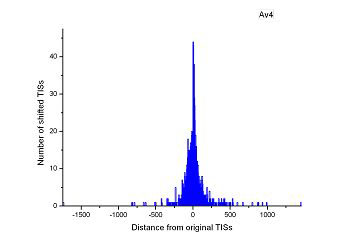 |
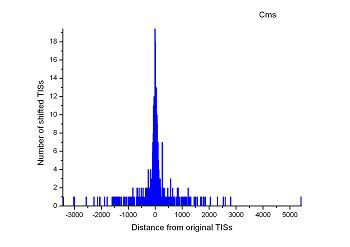 |
| (21) Agrobacterium vitis S4 | (22) Clavibacter michiganensis subsp. sepedonicus ATCC 33113 |
 |
 |
| (23) Candidatus Phytoplasma australiense | (24) Candidatus Phytoplasma mali |
 |
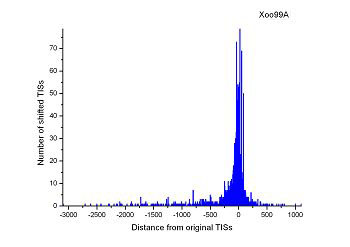 |
| (25) Xanthomonas campestris pv. campestris str. B100 | (26) Xanthomonas oryzae pv. oryzae PXO99A |
 |
 |
| (27) Xylella fastidiosa M12 | (28) Xylella fastidiosa M23 |