Welcome to ncRNA-eQTL
Numerous studies indicate that ncRNAs have critical functions across biological processes, and single nucleotide polymorphisms (SNPs)
could contribute to diseases or traits through influencing ncRNA expression. However, the associations between SNPs and ncRNA expressions are
largely unknown.
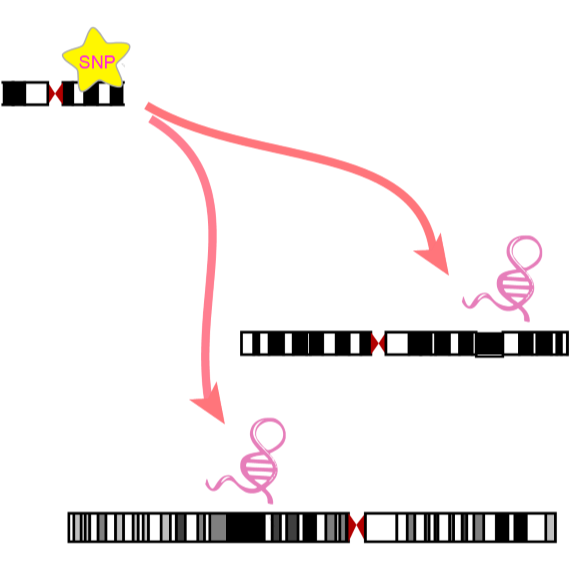
ncRNA-eQTL aims to comprehensively provide ncRNA related cis-eQTLs
(SNPs affect local ncRNA gene expression)
and trans-eQTLs (SNPs affect distant ncRNA gene expression) in different cancer types of The Cancer Genome Atlas
(TCGA). We have identified 6,045,445 and
715,952 eQTL−gene pairs at FDR < 0.05 in 33 cancer types in cis-eQTL and
trans-eQTL analysis, respectively. To prioritize promising ncRNA-eQTLs, we identified ncRNA-eQTLs
associated with
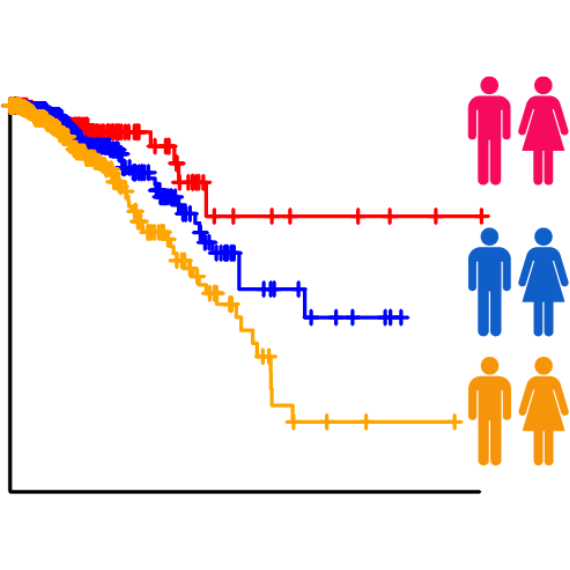
patient survival times (survival-eQTLs) and located in GWAS linkage disequilibrium (LD) regions (GWAS-eQTLs). In
survival-eQTL, we identified 8,235 eQTLs
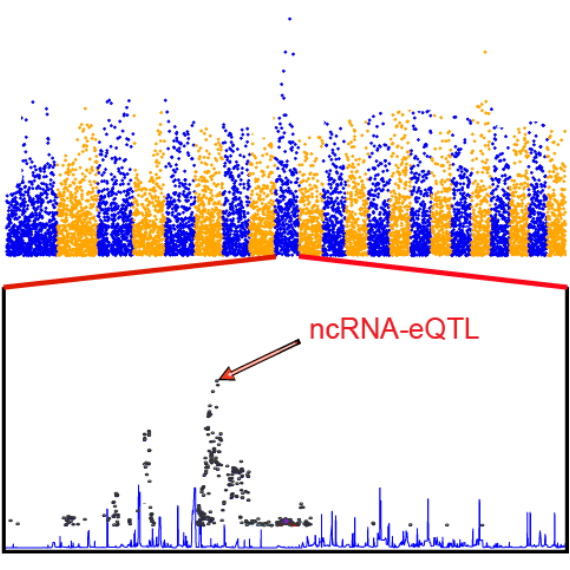
associated with patient overall survival times, and in GWAS-eQTL, we
identified 1,709,372 eQTLs that overlap with GWAS linkage disequilibrium (LD) regions.
ncRNAs in ncRNA-eQTL database includes lncRNAs (expression profiles obtained from RNA-seq) and miRNAs (expression profiles obtained from miRNA-seq). microRNA related eQTLs can be found at miRNA home.
Users can do:
Citation:
ncRNA-eQTL: a database to systematically evaluate the effects of SNPs on non-coding RNA expression across cancer types.Jiang Li, Yawen Xue, Muhammad Talal Amin, Yanbo Yang, Jiajun Yang, Wen Zhang, Wenqian Yang, Xiaohui Niu, Hong-Yu Zhang, Jing Gong*. Nucleic Acids Research.(2019)
e.g. SNP "rs11902447", Gene "NACA3P"