Definition
of Topological Parameters
Definition
of CO
Relative contact order is the average sequence distance between
all pairs of contacting residues normalized by the total sequence length:
![]() ,
,
where N is
the total number of contacts, ![]() is the sequence separation,
in residues, between contacting residues i and j, and L is the total
number of residues in the protein.1 As a measure of topological
complexity, CO reflects the relative importance of local and non-local contacts
to a protein’s native structure.
is the sequence separation,
in residues, between contacting residues i and j, and L is the total
number of residues in the protein.1 As a measure of topological
complexity, CO reflects the relative importance of local and non-local contacts
to a protein’s native structure.
Definition
of Abs_CO Absolute contact order is defined
as:
![]() .
.
Abs_CO takes into account the
influence of protein size on the folding rate.2
Definition
of LRO
Long range order for a protein is defined as:
![]() ,
,
where i and j are two residues for which the ![]() distance is
distance is ![]() 8 (Å) and N is the total number of residues in a
protein.3
8 (Å) and N is the total number of residues in a
protein.3
Definition
of FLOCAL As its name, fraction of
local contacts is defined as the fraction of local contacts in the total number
of contacts:
 ,
,
where ![]() if residues i and j are in contact, and 0 otherwise.4,5
FLOCAL is believed an
effective measure of helix/turn content and able to characterize the relative
importance of the local contacts in determining the folding rate of a protein.
if residues i and j are in contact, and 0 otherwise.4,5
FLOCAL is believed an
effective measure of helix/turn content and able to characterize the relative
importance of the local contacts in determining the folding rate of a protein.
Definition
of TCD
Total contact distance for a protein is defined as:
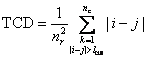 .
.
TCD is related to CO and LRO by a simple multiplication
(TCD = CO × LRO) if LRO is calculated with the same lcut value as CO.6
Definition
of CTP
Chain topology parameter is defined as:
![]() ,
,
where L is
the number of residues of the protein (chain length), N is the number of inter-residue contacts in the protein molecule,
and ![]() is the separation
in sequence between the contacting residues i and j.7 It is interesting to notice that the sequence
separation
is the separation
in sequence between the contacting residues i and j.7 It is interesting to notice that the sequence
separation ![]() has been squared
compared with CO.
has been squared
compared with CO.
Definition of LR_CO As a combination of relative contact order and long rage order, long range contact order for a protein takes the form of contact order:
![]()
but the "contact" is defined as in long range order.8
Definition
of CC
By treating protein structure as network of native contacts, clustering coefficient,
also termed as cliquishness, is defined for a given site (residue) i in the sequence
as:
 ,
,
where Nc is the number of
contacts to which site i
takes part.9
Definition of
Abs_CC By analogy with Abs_CO,
Abs_CC for a protein structure is defined here as:
![]() ,
,
where CC is the cliquishness for the structure.
Inputs:
- PDB file: the protein structure file in PDB format (usually with the extension of ".pdb" or ".ent"). Note that the PDB file can be provided in some compressed format (files with ".Z" and ".gz" extensions are acceptable).
- PDB code: the four lettered PDB ID. When it is provided, the PDB structure will be downloaded from the PDB database (http://www.pdb.org). It takes a while for the downloading process.
- Chain: For the structure with multiple chains, chain label should be indicated. Default is the first chain.
- Residue Range: a part of a chain can be designated by providing the start position i and the end position j, both i and j are integers and satisfying 0 < i < j < Length of the chain.
- Topological Parameters : indicating what parameter to be calculated. Totally, nine paramters can be calculated. For each parameter with optional thresholds of sequence separation (Lcut) and/or distance definition (Rcut) between two contacting residues, the thresholds are given the default values according to the corresponding original publication for that parameter. One can adjust these cut-off values for his own purpose.
Outputs:
The calculation results of the selected parameters for the submitted protein structure. Results will be returned on a new web page. If the optional email address is provided, it will also receive the results.
Warning:
Possible errors may occur when the PDB files contain broken chains or other defects. Null values may be returned for some parameters because of improper Lcut or Rcut setting. The email message may be blocked by the receiver (the server for the user's email address); if this happens, please try another email address.
References
1. Plaxco, K. W.,
Simons, K. T. & Baker, D. (1998). Contact order, transition state placement
and the refolding rates of single domain proteins. J Mol Biol 277, 985-94.
4. Mirny, L. & Shakhnovich, E.
(2001). Protein folding theory: from lattice to all-atom models. Annu Rev Biophys
Biomol Struct 30, 361-96.
5. Kuznetsov,
I. B. & Rackovsky, S. (2004). Class-specific correlations between
protein folding rate, structure-derived, and sequence-derived descriptors. Proteins 54, 333-41.
6. Zhou,
H. & Zhou, Y. (2002). Folding rate prediction using total
contact distance. Biophys
J 82, 458-63.
7. Nolting,
B., Schalike, W., Hampel, P., Grundig, F., Gantert, S., Sips, N., Bandlow, W.
& Qi, P. X. (2003). Structural determinants of the rate
of protein folding. J Theor
Biol 223,
299-307.
9. Micheletti, C. (2003). Prediction of
folding rates and transition-state placement from native-state geometry.
Proteins 51, 74-84.