Description
SBpred is a structure-based predictor of protein folding rate and folding type. SBpred predicts protein folding type according to the value (sign) of IFT, which was defined in our previous work [1] as follows:
![]()
Based on the training dataset, the coefficients in the above formula can be determined [1]. The prediction accuracy of folding type by IFT is as high as 85% (jack-knife test). See Ref [1] for details. The classification of the training data by IFT is illustrated in the figure below.
The folding rate is predicted by a newly defined indicator IRT, which is defined as follows:
![]() ,
,
where Length is the designated length of protein chain (Note: Length is default to be the chain length, but if the "Residue Range" is designated, the Length will be the length of the designated segment of the protein chain), Csum is the sum of contents of the amino acids that significantly correlate with the protein folding rates (On the training dataset, Csum is defined as Csum = (NC-NN-NQ+NT+NV)/Length, where NX (X = C, N, Q, T, V) are the occurrence numbers of the amino acids X, respectively), LR_CO is the long range contact order [1] (thresholds for "contact" definition are lcut = 2 and Rcut = 6) [1], and CC is the clustering coefficient (also called cliquishness) of the protein strucutre [2]. The coefficients (a, b, c, d, e) in IRT are determined based on the training dataset of 62 natural proteins [3]. The correlation between the predicted folding rates (indicated by IRT values) and the experimental folding rates is illustrated in the following figure.
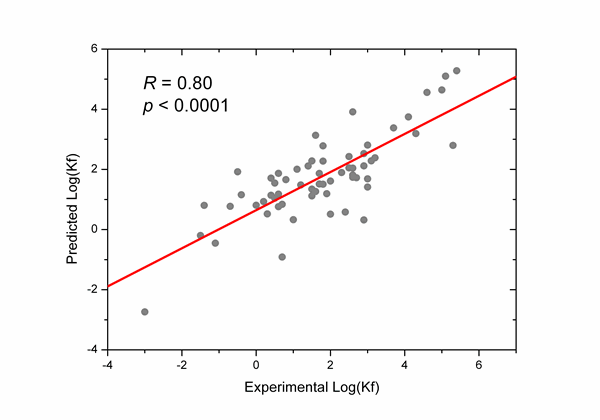
The correlation coefficient R between the predicted folding rates and the experimental folding rates in the "leave-one-out" jack-knife test is 0.76 (p < 0.0001).
Inputs:
Outputs:
The predicted folding rate and/or folding type of the provided protein structure. It will be returned on a new web page. If the optional email address is provided, it will also receive the results.
Warning:
Possible errors may occur when the PDB files contain broken chains or other defects. The email message may be blocked by the receiver (the server for the user's email address); if this happens, please try another email address.
References
[1] Bin-Guang Ma, Ling-Ling Chen, Hong-Yu Zhang. (2007) What determines protein folding type? An investigation of intrinsic structural properties and its implications for understanding folding mechanisms. Journal of Molecular Biology 370, 439-448. DOI: http://dx.doi.org/10.1016/j.jmb.2007.04.051.
[2] Micheletti, C. (2003). Prediction of folding rates and transition-state placement from native-state geometry. Proteins 51, 74-84.
[3] Ivankov DN and Finkelstein AV. (2004) Prediction of protein folding rates from the amino acid sequence-predicted secondary structure. PNAS 101, 8942-8944.