VMD Tutorial
VMD is designed for modeling, visualization, and analysis of biological systems such as proteins, nucleic acids, lipid bilayer assemblies, etc. It may be used to view more general molecules, as VMD can read standard Protein Data Bank (PDB) files and display the contained structure. VMD provides a wide variety of methods for rendering and coloring a molecule: simple points and lines, CPK spheres and cylinders, licorice bonds, backbone tubes and ribbons, cartoon drawings, and others. VMD can be used to animate and analyze the trajectory of a molecular dynamics (MD) simulation. In particular, VMD can act as a graphical front end for an external MD program by displaying and animating a molecule undergoing simulation on a remote computer.
Click here to learn more about VMD!
1. Load molecule
 |
 |
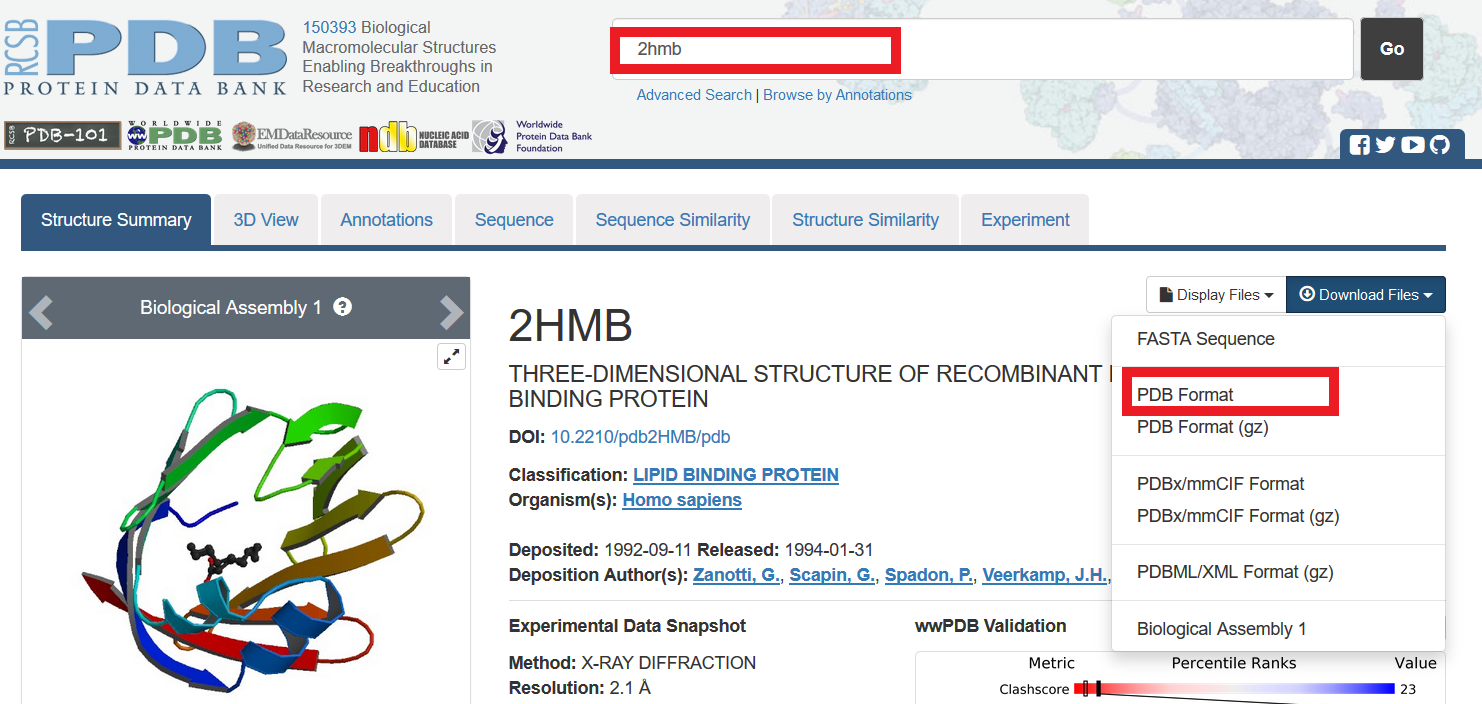
In "VMD Main" ,File -> NEW Molecule. In "Molecule File Browser",Browse -> select pdb file. In "Molecule File Browser", make sure "Determine file type" shows "PDB" from dropdown menu. It shouled be "PDB" automatically once you select a PDB file. |
|
In "Molecule File Browser",click Load. Now , the molecule display in "OpenGL Display"! |
2. Display protein
To see the three-dimensional structure of a protein, we use a variety of mouse modes. In "OpenGL Display" , Press move the mouse while holding the left mouse button, enter one step to observe what is happening. This is the mouse Rotation mode, through this mode, You can make this molecule around parallel axis rotation of the screen. If you press the right mouse button, Repeat the previous step, The numerator will rotate around an axis perpendicular to the screen |
|
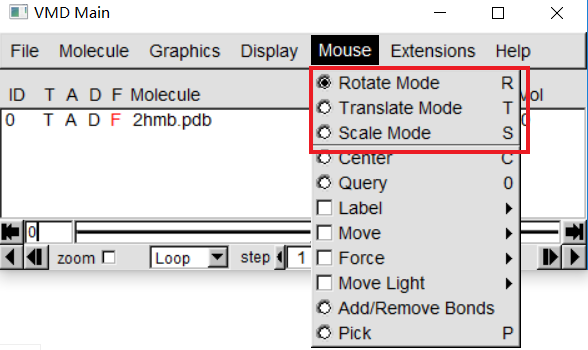 |
 In "VMD Main"-> Mouse, Here, you can change the mouse mode from "Rotation" to "Translation" or "Scale". Translation Mode allows you to hold down the left mouse button Key to move molecules on the screen. In Scale Mode, you can hold down the left button water move the mouse flat to zoom in or out. |
3. Visualization of the structure
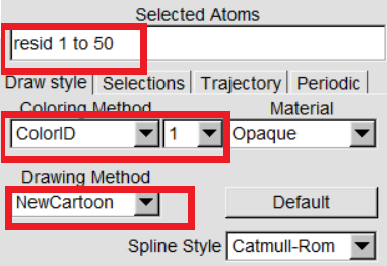
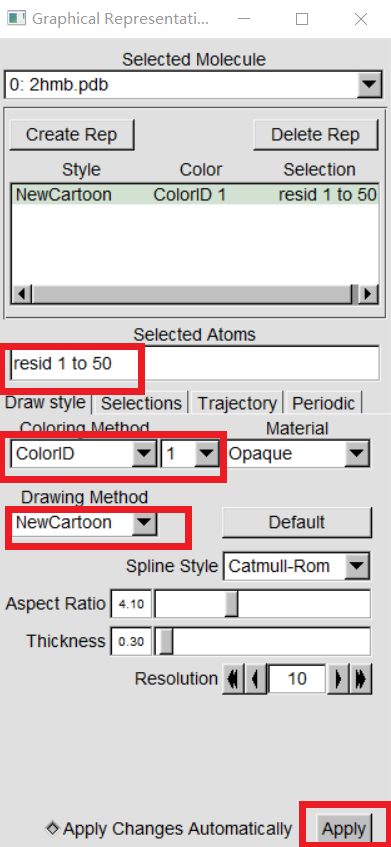
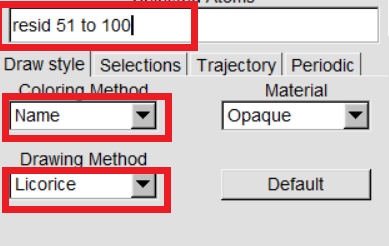
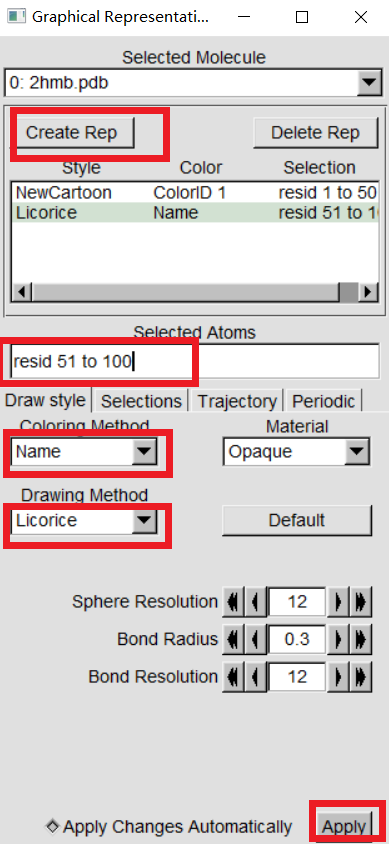
VMD can display your molecules in a variety of drawing modes. Here,we have to learn more about those that can help you drawing mode for different structures in proteins. In "VMD Main",Graphics -> Representations,we can see the graphical representation of the molecules currently displayed(lines). |
|
 |
 |
 |
 |
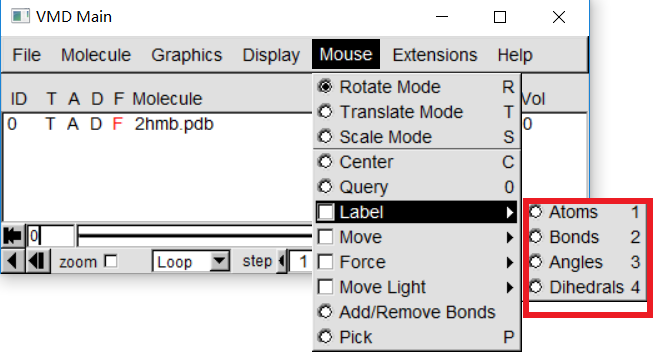
4. Obtain geometry information
 |
 |
 |
 |
 |
 |
For more information of your queried geometry, In "VMD Main", Graphics -> Labels. In the Labels menu, choose the category the drop down on the left corner. Then, click on the geometry from the box below, its information should be displayed.
5. Visualization window adjustment
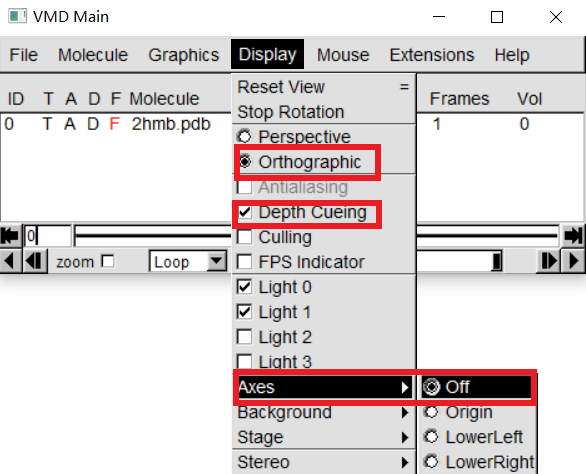 |
 In "VMD Main", Display -> Orthographic In "VMD Main", Display -> Axes -> Off In "VMD Main", Display -> Depth Cueing |
|
 |